Build & Running UDFs
An opinionated guide to making the most out of Fused UDFs
Fused UDFs are Python functions that run on serverless compute and can be called from anywhere with fused.run(udf). This guide is a resource meant to help you on your way to making the most out of UDFs.
A short reminder: The anatomy of a UDF
@fused.udf
def udf(my_awesome_input: int = 1):
import pandas as pd
return pd.DataFrame({"Look at my output: ": [my_awesome_input]})
Each UDF has a few specific elements:
- The
@fused.udfdecorator - Arguments -ideally typed-
- Imports inside the function
- Some logic
- A supported
returnobject
All of this is explained in the "Write UDF" section in much more details.
You can then run UDFs from anywhere with fused.run(udf). These are still Python functions, giving you a lot of flexibility on what oyu can do, but we have some recommendations for keeping them fast & efficient.
Writing efficient UDFs
Keep things small
The main benefit of Fused UDFs is how responsive they are. They achieve this by running on Python serverless compute. They can time out, so the best way to keep workflows fast is to keep them small:
- Break pipelines into single-task UDFs
- Leverage
fused.run()to chain UDFs together - Or run small tasks in parallel
Example: Breaking down a complex pipeline into smaller UDFs
❌ Not recommended:
@fused.udf
def inefficient_pipeline_udf(data_path):
import pandas as pd
df = pd.read_csv(data_path)
# Some complicated processing logic to create df_processed
processed_df = ...
return processed_df
✅ Instead, break it down:
@fused.udf
def load_data_udf(data_path):
import pandas as pd
return pd.read_csv(data_path)
@fused.udf
def process_data_udf(df):
import pandas as pd
# Some complicated processing logic to create df_processed
processed_df = ...
return processed_df
@fused.udf
def pipeline_udf(data_path):
import pandas as pd
df = fused.run(load_data_udf, data_path=data_path)
processed_df = fused.run(process_data_udf, df=df)
return processed_df
Run often, Iterate quickly
Just like writing short cells when developing in a Jupyter Notebook, we recommend you keep your UDFs short & fast to execute
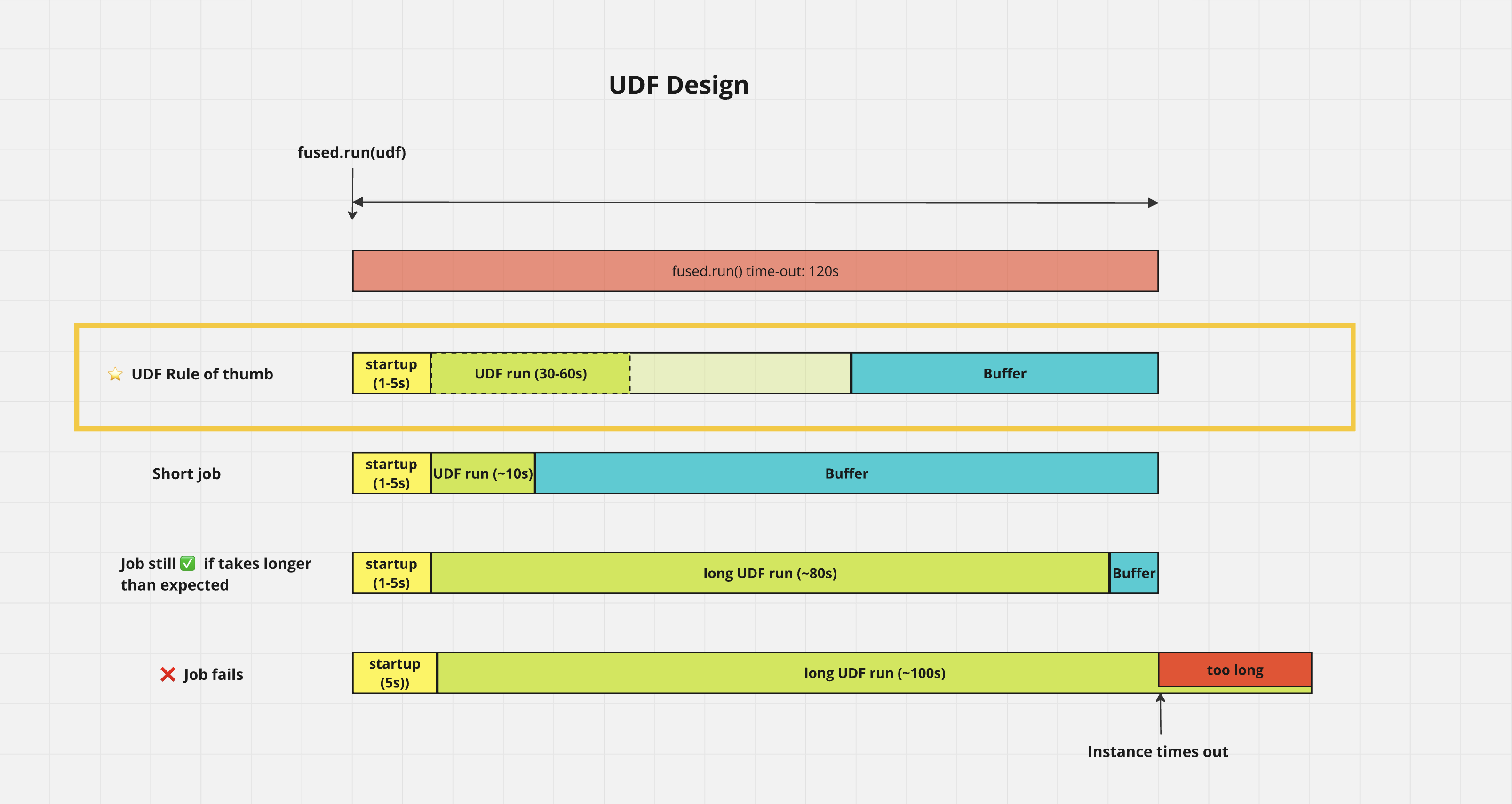
⚡️ Aim for UDFs that take up to 1min to run
UDFs run with fused.run() time out after 120s so we recommend you keep a buffer in case your UDF takes a bit longer to execute
Visual: UDF timing guideline
This is a breakdown of what happens when you run a UDF with fused.run() and why we recommend you keep your UDFs at the 30s-1min mark:
Run tasks in parallel
Sometimes you need to run a UDF over a large amount of inputs that takes longer than 120s to run, for example loading a large dataset or running a more complex process.
In this case you can use fused.submit() to run a small UDF over a set of inputs in parallel.
Example: Parallelizing a UDF with fused.submit()
Let's imagine we have a fetch_single_data that loads data from an API for a large amount of inputs input_data=[0,1,2,3,4,5,7,8,9]:
@fused.udf
def fetch_single_data(single_input: int):
import pandas as pd
import time
# Considering this as our API call, sleeping to simulate the time it takes to get the data
time.sleep(3)
return pd.DataFrame({"data": [f"processed_{str(single_input)}"]})
If we were to run this UDF in Workbench we would only be able to run it for 1 input at a time, so we could edit our UDF to loop over the inputs:
@fused.udf
def fetch_data(inputs: list):
import pandas as pd
import time
fetched_data = []
for i in inputs:
# Considering this as our API call
time.sleep(3)
fetched_data.append(f"processed_{str(i)}")
return pd.DataFrame({"data": fetched_data})
However, running this UDF with fused.run(fetch_data, inputs=input_data) will take longer as we add inputs, we could even quickly go over the 120s limit. We still do want to fetch data across all our inputs which is where fused.submit() comes in:
Going back to our original UDF, we can now run it with fused.submit() to run it in parallel:
@fused.udf
def load_data_udf(input_data):
results = fused.submit(
fetch_single_data,
input_data,
engine='local', # This ensures the UDF is run in our local server rather than spinning up new instances.
)
fetched_data = results.collect_df()
return fetched_data
This is of course a simplified example, but it shows how you can use fused.submit() to run a UDF in parallel for each input.
This now runs a lot faster by running the fetch_single_data UDF in parallel for each input.
The example here blocks the main thread until all the fused.submit() calls have finished. This means you might have to wait longer in Workbench for the results to show up.
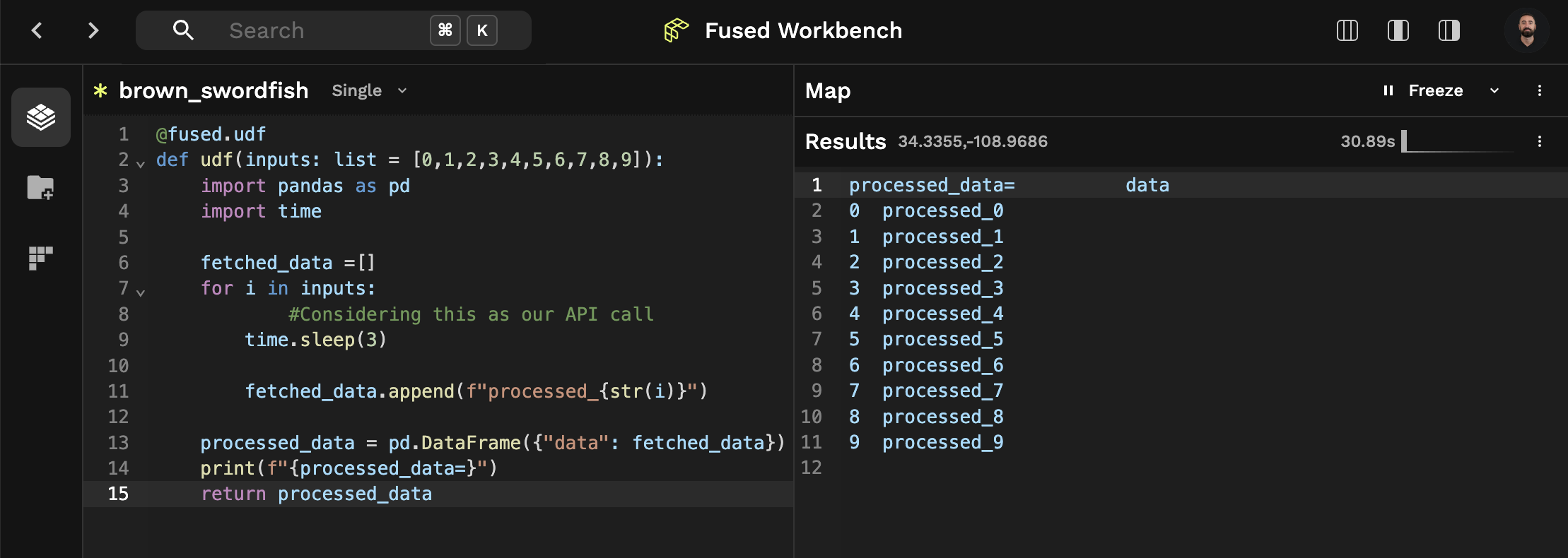
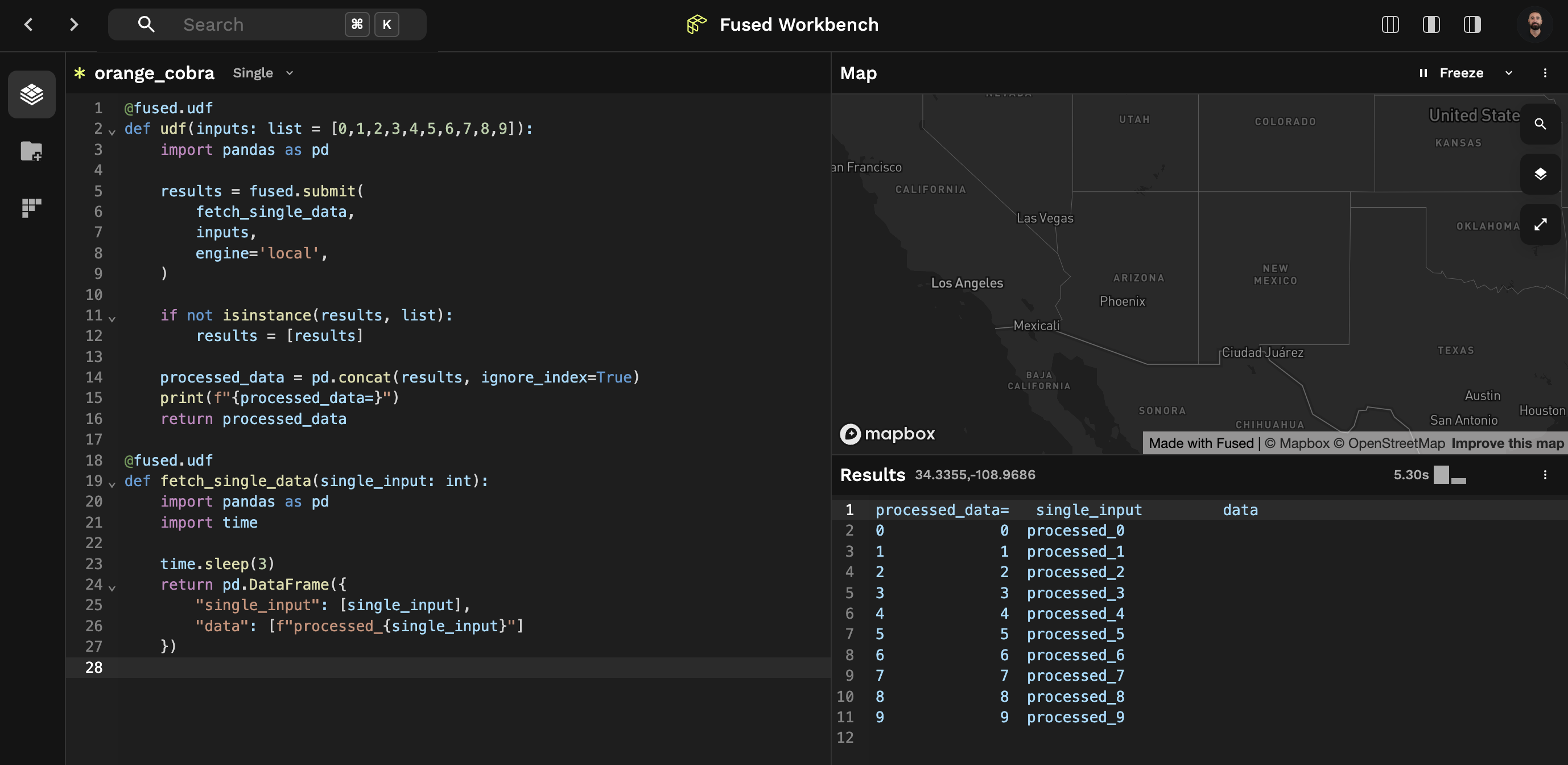
Comparison of both approaches in Workbench:
Running with fused.run(), 30.89s:
Running with fused.submit(), 5.3s:
Use offline instances for your large jobs
You might not be able to run your UDF within the 120s limit of the default fused.run(udf) call or break it down into smaller UDFs and parallelize it with fused.submit(). In that case you can use offline instances. These also allow you to:
- Choose instances with more memory & CPU
- Run your UDF for longer than 120s (at the expense of slower startup time and requiring to save your data to storage to retrieve it)
You can find an example of using offline instances in one of our end to end examples when ingesting data into portioned cloud native formats
Cache as much as you can
Fused relies heavily on caching repetitive tasks to make recurring calls much faster (and more compute efficient)
✅ You want to use caching for functions with inputs that are recurring:
- Loading a dataset
- Computing a recurring operation with default variables
- Intermediate results you'll reuse soon
❌ When not to use caching:
- In most cases, for functions taking
boundsas an argument -> your function + input cache would get re-generated for each newbounds(which changes each time you pan around in Workbench Map view for example) - Data you want others in your team or external to Fused to use. You're better off writing your data to cloud storage like
s3orgcs
Example: Caching a repetitive task
Re-using the example from keeping things small:
❌ Not recommended:
@fused.udf
def inefficient_pipeline_udf(data_path):
import pandas as pd
df = pd.read_csv(data_path)
# Some complicated processing logic to create df_processed
processed_df = ...
return processed_df
✅ Instead, break it down AND cache the calls:
@fused.udf
def load_data_udf(data_path):
import pandas as pd
return pd.read_csv(data_path)
@fused.udf
def process_data_udf(df):
import pandas as pd
# Some complicated processing logic to create df_processed
# ...
return processed_df
@fused.udf
def pipeline_udf(data_path):
import pandas as pd
@fused.cache
def load_data(data_path):
return fused.run(load_data_udf, data_path=data_path)
@fused.cache
def process_data(df):
return fused.run(process_data_udf, df=df)
df = load_data(data_path)
processed_df = process_data(df)
return processed_df
Read more about the caching details:
- in the dedicated section
- How you can use cache to speed up exploration of slow to read datasets
Prepare your large datasets
Fused works at its best with data that is fast to read and can be read in tiles or chunks. We know that most of the data out there isn't in the most efficient file formats which is why we provide tools to ingest your own data into cloud-optimized, partitioned formats.
We have a dedicated page for when you should consider ingesting your own data. As a rule of thumb you want to consider ingesting your data when:
- Files are read multiple times and >100MB
- Files that are slow or require some processing to open (
.zipfor example)
Don't start from scratch: UDF Catalog
Just like using libraries in Python to leverage existing tools, you don't need to start from scratch in Fused. We have a Catalog of existing UDFs built & maintained by us and the community.
You can find a host of different UDFs that can serve as a starting point or as inspiration to create your own UDFs:
- Open datasets from common open repository like STAC catalogs
- Run on the fly ML prediction on satellite images
- Compute a spatial join between 2 datasets
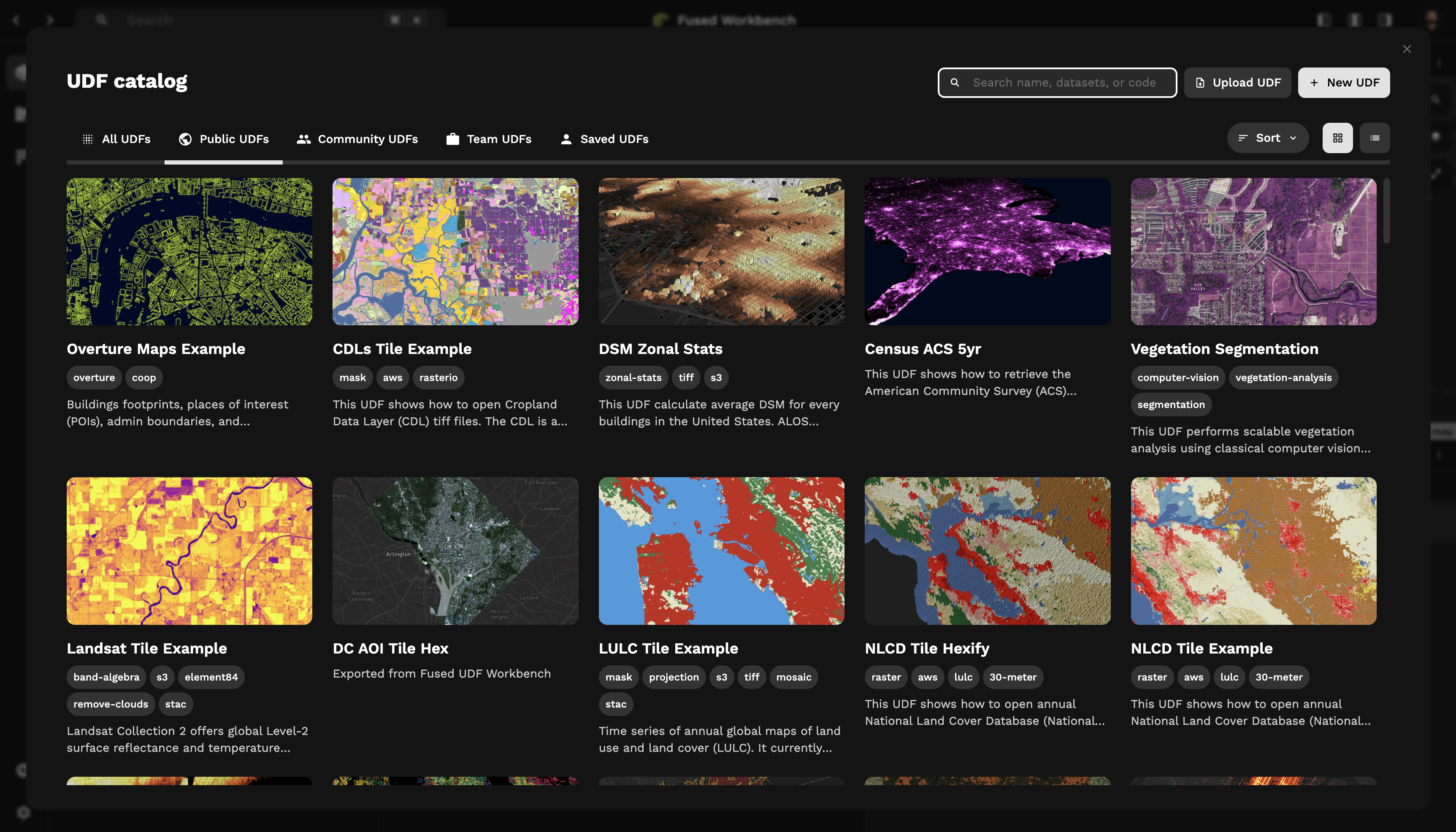
You can also contribute your own UDFs to the community!
Debugging UDFs
The reality of writing code is that stuff breaks, often and sometimes in mysterious ways. Here's some of our recommendations for how to debug your UDFs
Use print()
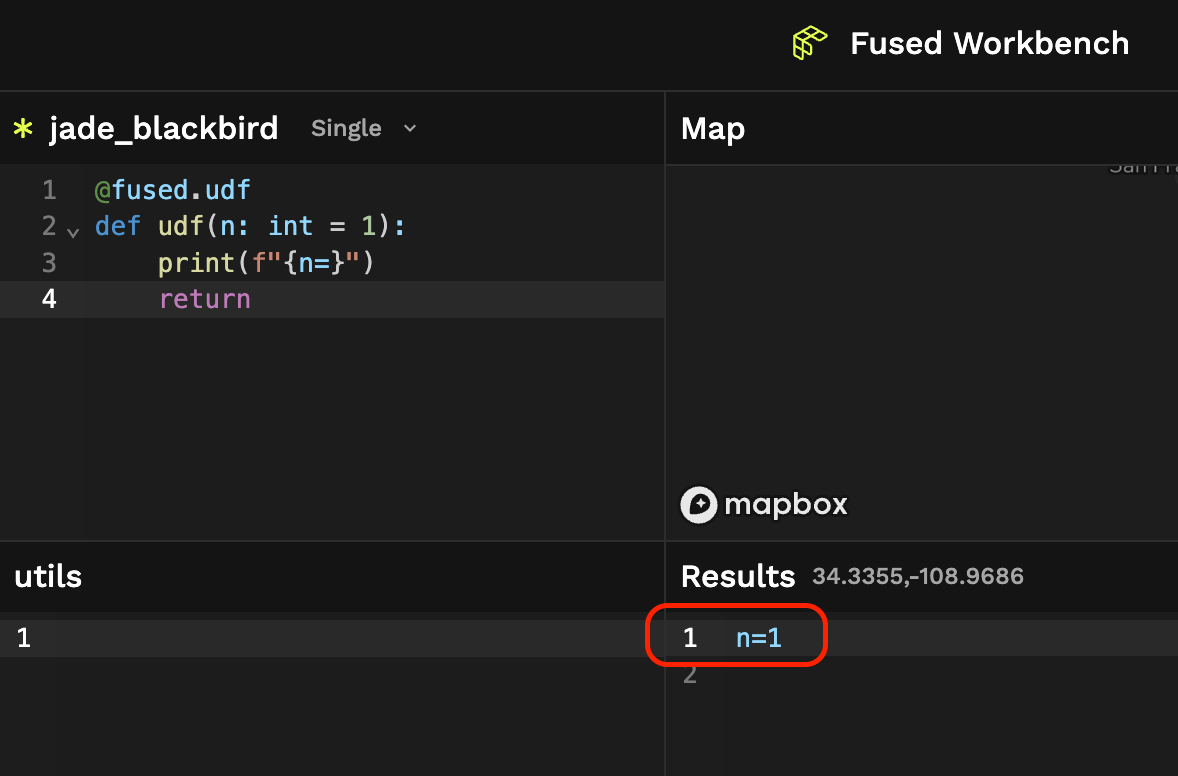
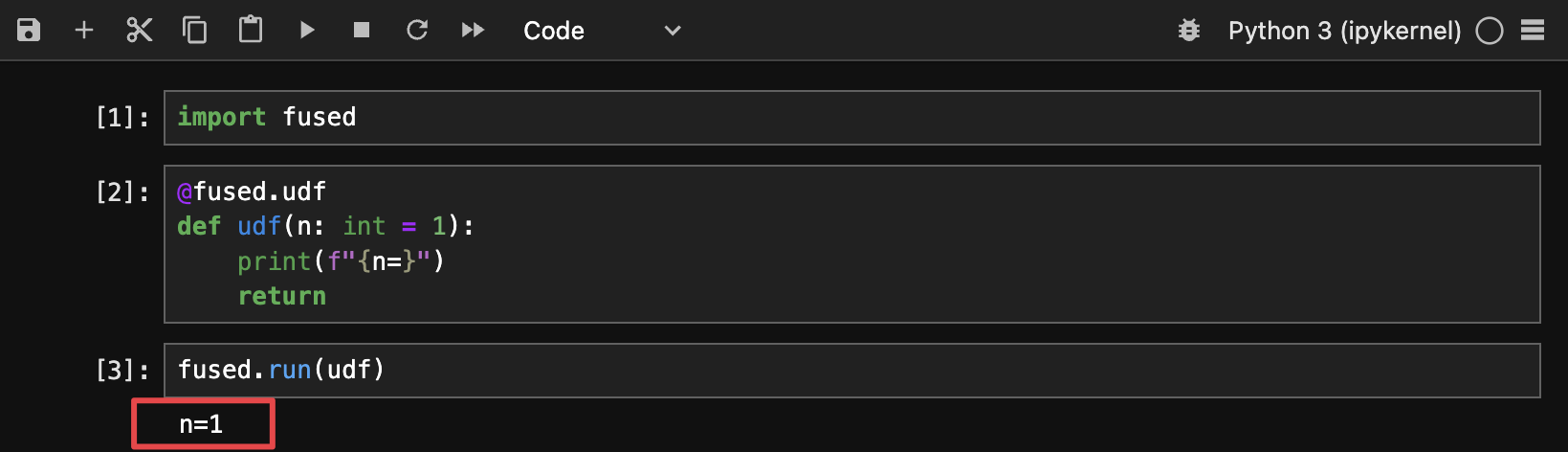
UDFs return stdout either in Workbench Code Editor or locally when running fused.run(udf) so the easiest way to get info about your UDFs is to use good old print:
@fused.udf
def udf(n: int = 1):
print(f"{n=}")
return
Since Python 3.8 you can use f-string debugging which is what we recommend you use:
print(f"{my_fancy_variable=}")
This allows you to print many variables without getting lost with what is what
- Workbench
- notebook
Type all your inputs
We strongly recommend you type all your inputs with the appropriate type:
@fused.udf
def udf(
bounds:fused.types.Bounds=None, n:int=1
):
...
return
This has 2 effects:
- It makes your code more readable to others
- Fused only supports a few types at the moment. Any non-typed or unsupported types will be passed as
str
Use time.time()
Sometimes you're not sure what's taking so long. The simplest way to figure this out is to use time.time():
Example: finding a slow process
@fused.udf
def udf():
import time
beginning_time = time.time()
# long processing step #1
time.sleep(5)
end_process_1 = time.time()
process_time_1 = round(
end_process_1 - beginning_time, 2
)
print(f"{process_time_1=}")
# short processing step
time.sleep(0.2)
process_time_2 = round(
time.time() - end_process_1, 2
)
print(f"{process_time_2=}")
return
Would give us:
>>> process_time_1=5.0
>>> process_time_2=0.2
Test out your UDFs before running them in parallel
When using fused.submit() to run a UDF in parallel you can use the debug_mode to run the 1st argument directly to test if your UDF is working as expected:
job = fused.submit(udf, inputs, debug_mode=True)
This runs fused.run(udf, inputs[0]) and returns the results. It allows you to quickly test out udf before running it on a large number of inputs.
Since UDFs are cached by default, using fused.submit(udf, inputs, debug_mode=True) means Fused won't run the 1st input again, as you'll have just run it! (unless udf is set with cache_max_age=0)
Join the Discord for support
We host & run a Discord server where you can ask any questions! We or the community will do our best to help you out!