Data Ingestion
This page will give you all the tools to make your data fast to read to make your UDFs more responsive.
What is this page about?
The whole purpose of Fused is to speed up data science pipelines. To make this happen we need the data we're working with to be responsive, regardless of the dataset. The ideal solution is to have all of our data sitting in RAM right next to our compute, but in real-world applications:
- Datasets (especially geospatial data) can be in the Tb or Pb range which rarely fit in storage, let alone RAM
- Compute needs to be scaled up and down depending on workloads.
One solution to this is to build data around Cloud Optimized formats: Data lives in the cloud but also leverages file formats that are fast to access. Just putting a .zip file that needs to be uncompressed at every read on an S3 bucket is still very slow. Our ingested data should be:
- On the cloud so dataset size doesn't matter (AWS S3, Google Cloud Storage, etc.)
- Partitioned (broken down into smaller pieces that are fast to retrieve so we can load only sections of the dataset we need)
This makes it fast to read for any UDF (and any other cloud operation), so developing UDFs in Workbench UDF Builder & running UDFs is a lot faster & responsive!
Benchmark & Demo
We're going to use a real-world example to show the impact of using different file formats & partitioning to make data a lot faster to access. For this demo, we'll be using AIS (Automatic Identification System) data as for our Dark Vessel Detection example. These are points which represent the location of boats at any given time. We'll be using free & open data from NOAA Digital Coast.
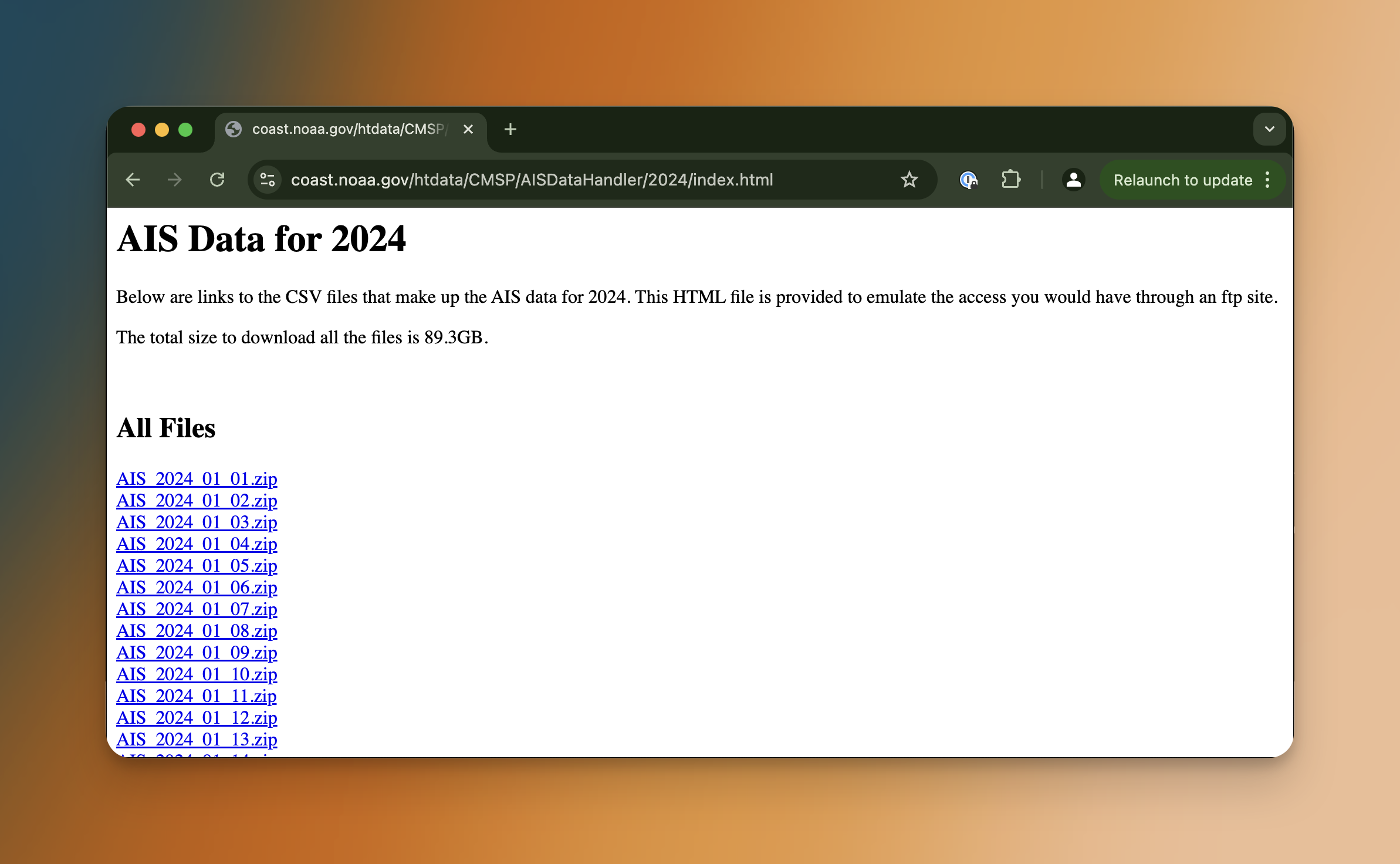
The NOAA Digital Coast platform gives us 1 zip file per day with the location of every boat with an AIS transponder as CSV (once unzipped).
We'll download 1 day and upload it as a CSV to Fused server with fused.upload():
@fused.udf
def save_ais_csv_to_fused_udf():
"""Downloading a single day of data to Fused server"""
import requests
import zipfile
import pandas as pd
response = requests.get("https://coast.noaa.gov/htdata/CMSP/AISDataHandler/2024/AIS_2024_01_01.zip")
with open("data.zip", "wb") as f:
f.write(response.content)
with zipfile.ZipFile("data.zip", "r") as zip_ref:
zip_ref.extractall("./data")
csv_df = fused.api.upload("./data/AIS_2024_01_01.csv", "fd://demo_reading_ais/AIS_2024_01_01.csv")
print(f"Saved data to fd://demo_reading_ais/")
return pd.DataFrame({"status": 'Done'})
And simply running this UDF:
fused.run(save_ais_csv_to_fused_udf)
We can check that our CSV was properly ingested with File Explorer by navigating to fd://demo_reading_ais/:
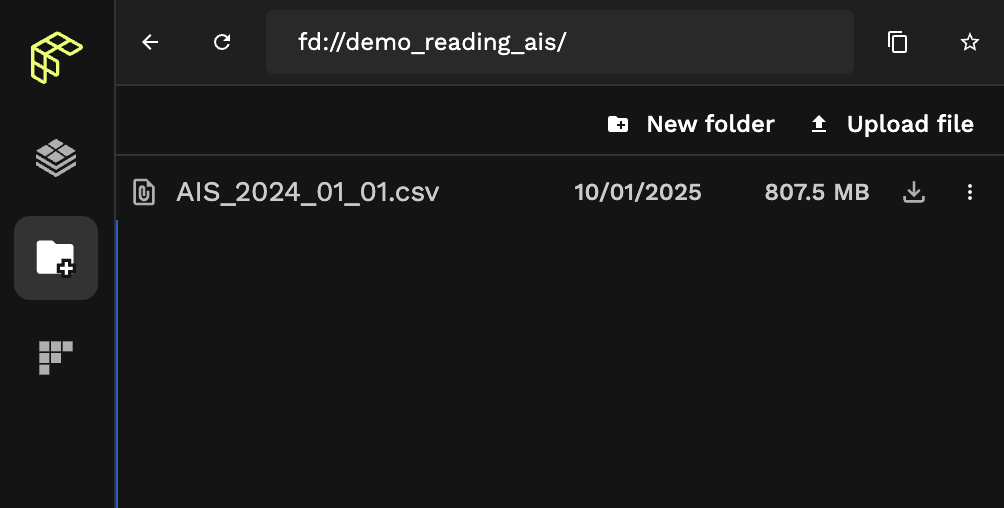
That's one big CSV.
But opening it on its own doesn't do all that much for us. We're going to create 3 UDFs to showcase a more real-world application: Opening the dataset and returning a subset inside a bounding box. We'll do this 3 different ways to compare their execution time:
-
- From the default CSV
-
- From the same data but saved a
.parquet
- From the same data but saved a
-
- Ingesting this data with
fused.ingest()and reading it from our ingested data
- Ingesting this data with
Since our AIS data covers the waters around the US, we'll use a simple bounding box covering a small portion of water:
import shapely
bbox = gpd.GeoDataFrame(geometry=[shapely.box(-81.47717632893792,30.46235012285108,-81.33723531132267,30.58447317149745)])
This bbox is purposefully small (print(smaller_bbox.iloc[0].geometry.area) returns 0.02) to highlight loading a very large dataset and recover only a small portion of data.
1. Reading directly from CSV
Here's a simple UDF to read our CSV in memory and return only points in within our bounding box:
@fused.udf
def from_csv_df_udf(
bbox: fused.types.ViewportGDF,
path: str="fd://demo_reading_ais/AIS_2024_01_01.csv"
):
import geopandas as gpd
import pandas as pd
df = pd.read_csv(path)
gdf = gpd.GeoDataFrame(df, geometry=gpd.points_from_xy(df.LON, df.LAT))
bbox_ais = gdf[gdf.geometry.within(bbox.iloc[0].geometry)]
return bbox_ais
2. Reading from Parquet
First we need to save our AIS data a .parquet:
@fused.udf
def ais_to_parquet():
import pandas as pd
# S3 bucket & dir for demo purpose here. Replace with your own
csv_df = pd.read_csv("s3://your-bucket/your-dir/demo_reading_ais/AIS_2024_01_01.csv")
csv_df.to_parquet("s3://your-bucket/your-dir/demo_reading_ais/AIS_2024_01_01.parquet")
print(f"Saved data (as parquet) to fd://demo_reading_ais/")
return pd.DataFrame({"status": ['Done']})
Here's our updated UDF to read a .parquet file:
@fused.udf
def from_parquet_udf(
bbox: fused.types.ViewportGDF,
path: str="s3://your-bucket/your-dir/demo_reading_ais/AIS_2024_01_01.parquet"
):
import geopandas as gpd
import pandas as pd
df = pd.read_parquet(path)
gpd = gpd.GeoDataFrame(df, geometry=gpd.points_from_xy(df.LON, df.LAT))
bbox_ais = gpd[gpd.geometry.within(bbox.iloc[0].geometry)]
return bbox_ais
3. Reading from Fused Ingested GeoParquet
Now, we're going to ingest the parquet file we have to create geo-partitioned files, using fused.ingest(). We'll go in more details on how to ingest your own data in the following section.
job = fused.ingest(
"fd://demo_reading_ais/AIS_2024_01_01.parquet",
"fd://demo_reading_ais/ingested/",
target_num_chunks=500, # 500 is a rough default to use in most cases. We're not optimizing this value for now
lonlat_cols=('LON','LAT')
)
Ingestion jobs are quite memory hungry so we'll run this on a remote machine as a large run:
job.run_remote(
instance_type='r5.8xlarge', # 256GiB RAM machine
)
Again using File Explorer to inspect our data we can see fused.ingest() didn't create 1 file but rather multiple:
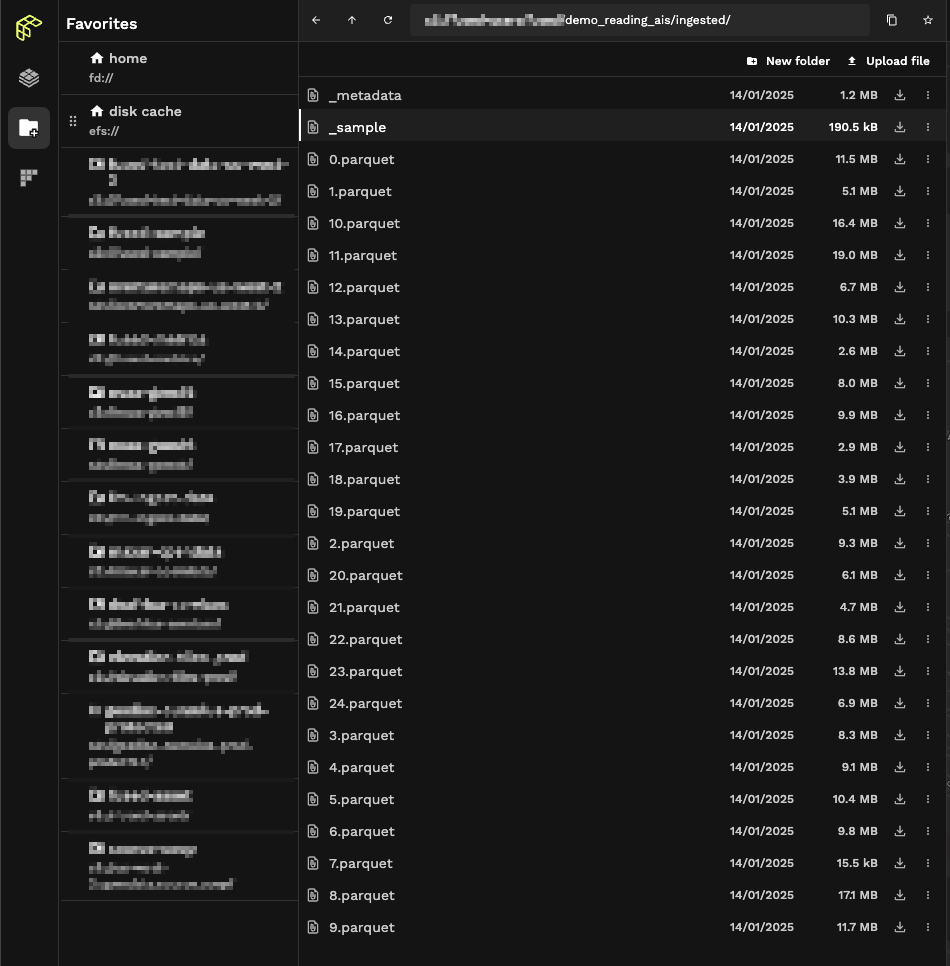
Our ingestion process broke down our dataset into smaller files (making each file easier to access) and a _sample.parquet file containing the bounding box of each individual file. This allows us to first intersect our bbox with _sample and then only open the smaller .parquet files we need:
@fused.udf
def read_ingested_parquet_udf(
bbox: fused.types.TileGDF,
path: str = "s3://your-bucket/your-dir/file_format_demo/ingested/",
):
import fused
import pandas as pd
# Build in fused method to reach the `_sample` file and return only bounding box of each parquet holding our points
df = fused.get_chunks_metadata(path)
# Only keeping the tiles where our bbox is -> Only need to load actual data inside / touching our bbox
df = df[df.intersects(bbox.geometry.iloc[0])]
# This is based on Fused's ingestion process
chunk_values = df[["file_id", "chunk_id"]].values
# We only need to now load the parquet files that were kept that touched our bbox
rows_df = pd.concat([
fused.get_chunk_from_table(
path, fc[0], fc[1], columns=['geometry']
) for fc in chunk_values
])
df = rows_df[rows_df.intersects(bbox.geometry[0])]
df.crs = bbox.crs
return df
We've implemented a utils function that allows you to more simply read Fused ingested data: table_to_tile
So instead of re-implementing the above in 2 lines of code you can read your ingested data:
@fused.udf
def udf(
bbox: fused.types.TileGDF, path: str='s3://your-bucket/your-dir/demo_reading_ais/ingested/'):
utils = fused.load('https://github.com/fusedio/udfs/tree/19b5240/public/common/').utils
df = utils.table_to_tile(bbox, table=path)
return df
Comparing all 3 runs
We'll run each of these 3 methods in a Jupyter notebook using the %%time magic command to compare their run time:
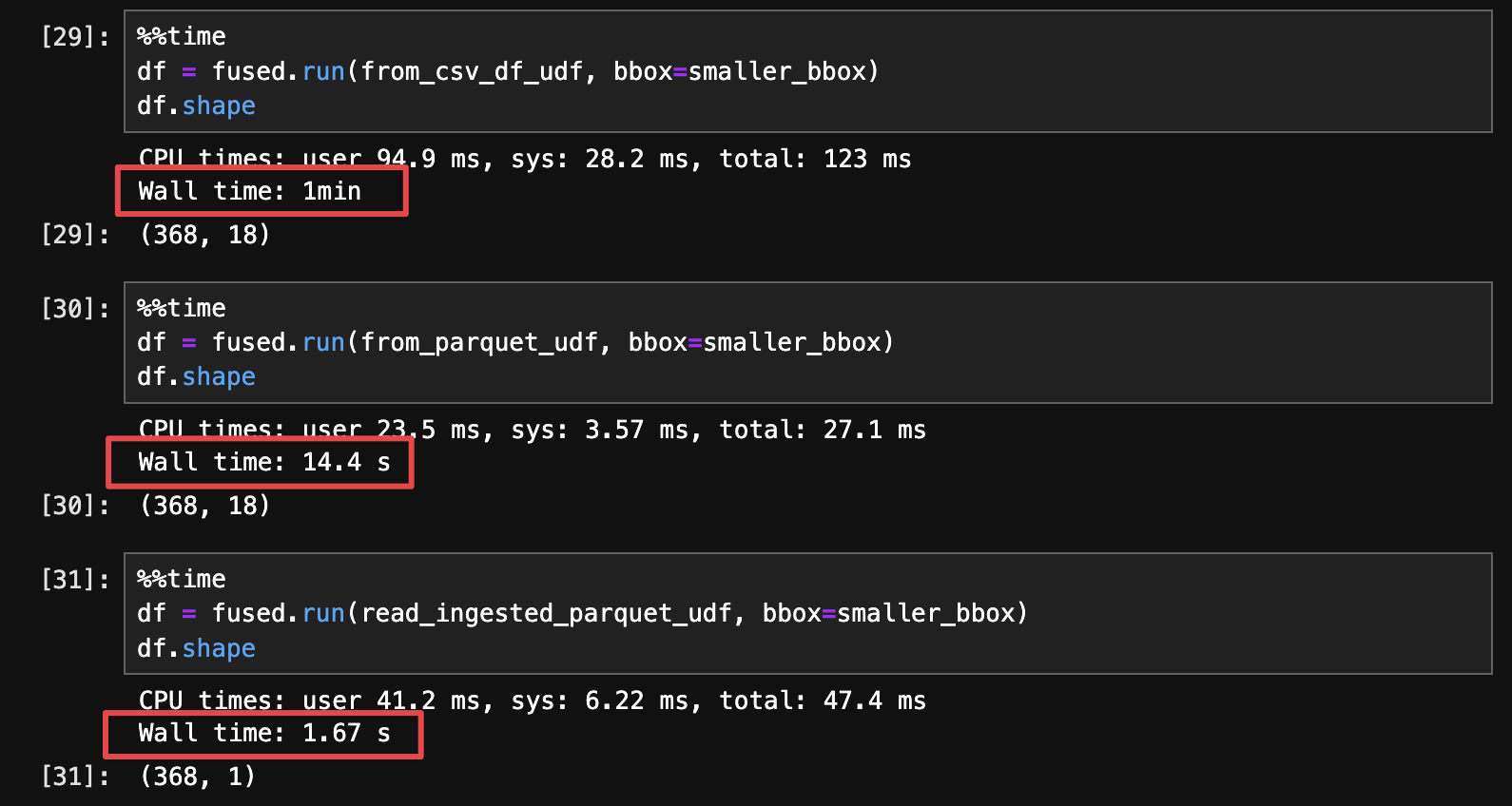
There are a few conclusions to draw here:
- simply saving a CSV to
.parquetmakes files smaller & significantly faster to read just by itself (4x speed gain in this example) - But proper geo-partitioning takes those gains ever higher (additional 8x speed gain in this specific example)
In short, by ingesting our data with fused.ingest() we're trading some up-front processing time for much faster read time. We only need to ingest our data once and every subsequent read will be fast and responsive.
When is ingestion needed?
You don't always need to ingest your file into a cloud, geo-partitioned format. There are a few situation when it might be simpler & faster to just load your data.
Small files (< 100Mb ) that are fast to open (already in .parquet for example) that you only read once (note that it might be read 1x in your UDF but your UDF might be run many times)
Example of data you should ingest: 1Gb .zip of shapefile
.zipmeans you need to unzip your file each time you open it and then read it. This slows down working with the data every minute. This results in each individual files (a CSV when unzipped) containing millions of points.- shapefile contains multiple files, it isn't the fastest to read
Example of data you don't need to ingest: 50Mb .parquet
- Even if the data isn't geo-partitioned, loading this data should be fast enough to make any UDF fast
Using cache as a single-use "ingester"
We could actually significantly speed up the above example where we loaded the AIS data as a CSV without running fused.ingest(), by using cache:
@fused.udf
def from_csv_df_udf(
bbox: fused.types.ViewportGDF,
path: str="s3://your-bucket/your-dir/demo_reading_ais/AIS_2024_01_01.csv"
):
import geopandas as gpd
import pandas as pd
@fused.cache
def load_csv(path):
import pandas as pd
return pd.read_csv(path)
df = load_csv(path)
gdf = gpd.GeoDataFrame(df, geometry=gpd.points_from_xy(df.LON, df.LAT))
bbox_ais = gdf[gdf.geometry.within(bbox.iloc[0].geometry)]
return bbox_ais
The first run of this would still be very slow, but running this a second time would give us a much faster result:
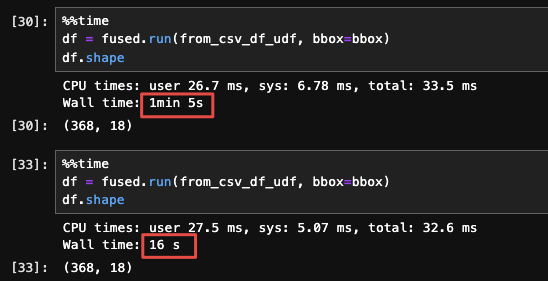
We're using @fused.cache to cache the result of load_csv() which is the same regardless of our bbox, so this allows us to save an 'intermediate' result on disk.
There are some limitations to this approach though:
- This cache is emptied after 24h.
- This cache is overwritten any time you change the cached function or its inputs
This approach is only helpful if you want to 1 time explore a new dataset in UDF Builder and don't want to wait around for the ingestion run to be done. Beyond that, this will end up being slower (and more frustrating)
When is ingestion needed?
You don't always need to ingest your file into a cloud, geo-partitioned format. There are a few situation when it might be simpler & faster to just load your data.
Small files (< 100Mb ) that are fast to open (already in .parquet for example) that you only read once (note that it might be read 1x in your UDF but your UDF might be run many times)
Example of data you should ingest: 1Gb .zip of shapefile
.zipmeans you need to unzip your file each time you open it and then read it. This slows down working with the data- shapefile contains multiple files, it isn't the fastest to read
Example of data you don't need to ingest: 50Mb .parquet
- Even if the data isn't geo-partitioned, loading this data should be fast enough to make any UDF fast
File Formats
For rasters (images)
For images (like satellite images) we recommend using Cloud Optimized GeoTiffs (COGs). To paraphrase the Cloud Native Geo guide on them:
Cloud-Optimized GeoTIFF (COG), a raster format, is a variant of the TIFF image format that specifies a particular layout of internal data in the GeoTIFF specification to allow for optimized (subsetted or aggregated) access over a network for display or data reading
Fused does not (yet) have a build-in tool to ingest raster data. We suggest you create COGs yourself, for example by using gdal's built-in options or cogger
Cloud Optimized GeoTiffs have multiple different features making them particularly interesting for cloud native applications, namely:
- Tiling: Images are split into smaller tiles that can be individually accessed, making getting only parts of data a lot faster.
- Overviews: Pre-rendered images of lower zoom levels of images. This makes displaying images at different zoom levels a lot faster
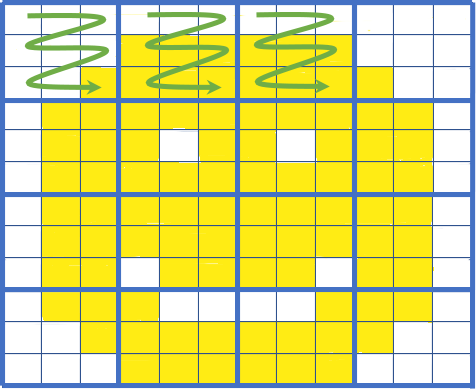
A simple visual of COG tiling: If we only need the top left part of the image we can fetch only those tiles (green arrows). Image courtesy of Element 84's blog on COGs
- Element84 wrote a simple explainer of what Cloud Optimized GeoTiffs are with great visuals
- Cloud Optimized Geotiff spec dedicated website
- Cloud Optimized Geotiff page on Cloud Native Geo guide
For vectors (tables)
To handle vector data such as pandas DataFrames or geopandas GeoDataFrames we recommend using GeoParquet files. To (once again) paraphrase the Cloud Native Geo guide:
GeoParquet is an encoding for how to store geospatial vector data (point, lines, polygons) in Apache Parquet, a popular columnar storage format for tabular data.
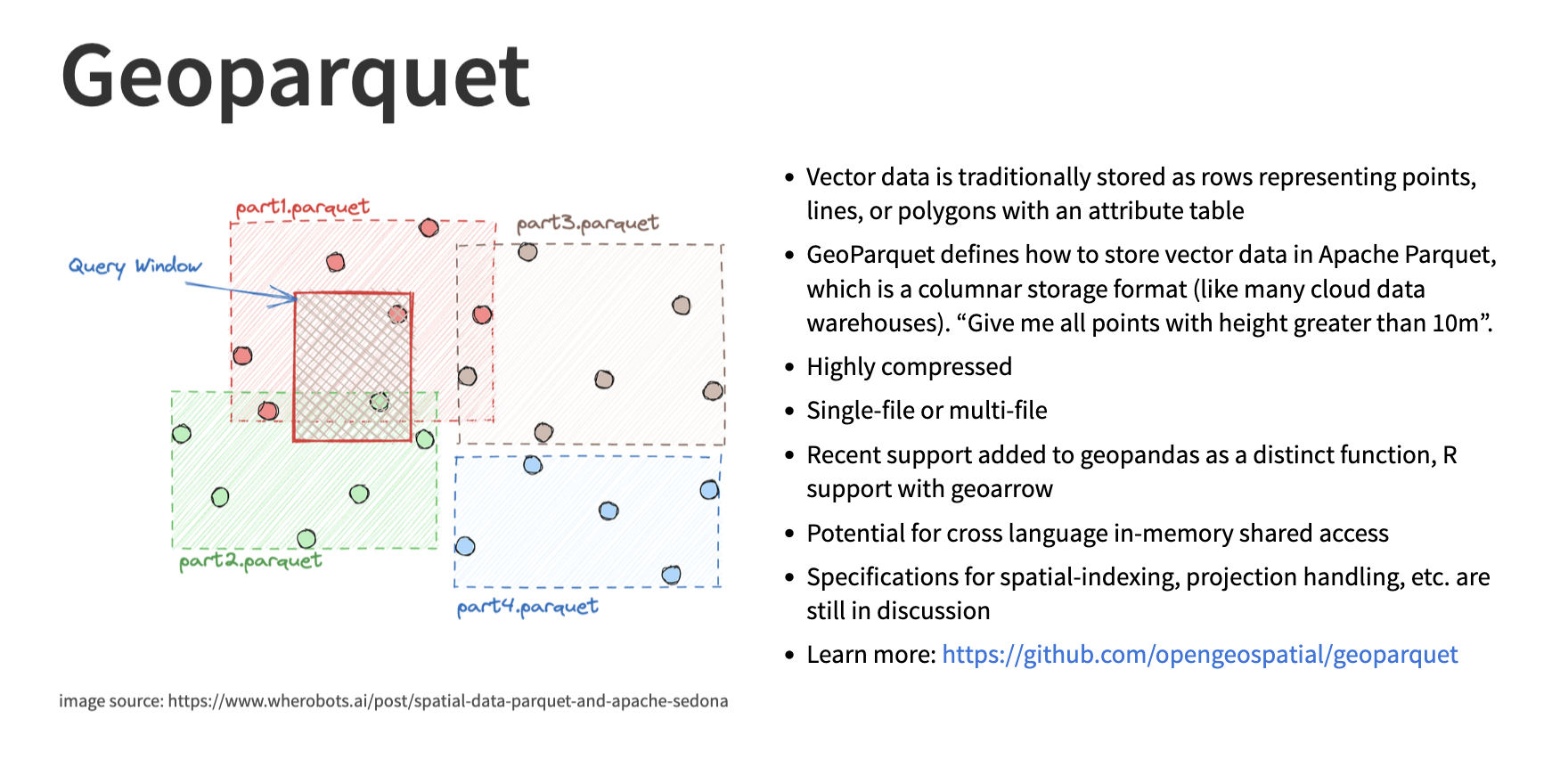
Image credit from the Cloud Native Geo slideshow
Refer to the next section to see all the details of how to ingest your data with Fused's built-in fused.ingest() to make the most out of geoparquet
Additional resources
- Read the Cloud-Optimized Geospatial Formats Guide written by the Cloud Native Geo Org about why we need Cloud Native formats
- Friend of Fused Kyle Barron did an interview about Cloud Native Geospatial Formats. Kyle provides simple introductions to some cloud native formats like
GeoParquet