Ingest your own data
This guide explains how to use fused.ingest to geopartition and load vector tables into an S3 bucket so they can quickly be queried with Fused.
Ingest geospatial table data
To run an ingestion job on vector data we need:
- Input data - This is could be CSV files, a
.zipcontaining shapely files or any other sort of non partitioned data - A cloud directory - This is where we will save our ingested data and later access it through UDFs
We've built our own ingestion pipeline at Fused that partitions data based on dataset size & location. Our ingestion process:
- Uploads the
input - Creates geo-partitions of the input data
This is defined with fused.ingest():
# Run this locally - not in Workbench
job = fused.ingest(
input="https://www2.census.gov/geo/tiger/TIGER_RD18/LAYER/TRACT/tl_rd22_11_tract.zip",
output=f"fd://census/dc_tract/",
)
Ingestion jobs often take more than a few seconds and require a lot of RAM (we open the whole dataset & re-partition it), which makes this a large run so we're going to use run_batch() so our ingestion job can take as long as needed.
To start an ingestion job, call run_batch on the job object returned by fused.ingest.
# Run this locally - not in Workbench
job_id = job.run_batch()
Refer to the dedicated documentation page for fused.ingest() for more details on all the parameters
Ingested tables can easily be read with the Fused utility function table_to_tile, which spatially filters the dataset and reads only the chunks within a specified polygon.
@fused.udf
def udf(
bounds: fused.types.Bounds = None,
table="s3://fused-asset/infra/building_msft_us/"
):
common = fused.load("https://github.com/fusedio/udfs/tree/fbf5682/public/common/")
return common.table_to_tile(bounds, table)
The following sections cover common ingestion implementations. You can run ingestion jobs from a Python Notebook or this web app.
Ingest a table from a URL
Ingests a table from a URL and writes it to an S3 bucket specified with fd://.
import fused
job_id = fused.ingest(
input="https://www2.census.gov/geo/tiger/TIGER_RD18/LAYER/TRACT/tl_rd22_11_tract.zip",
output=f"fd://census/dc_tract/",
).run_batch()
Get access to the logs:
job_id
If you encounter the message
HTTPError: {'detail': 'Quota limit: Number of running instances'}, please contact Fused to increase the number of workers in your account.
Ingest multiple files
import fused
job_id = fused.ingest(
input=["s3://my-bucket/file1.parquet", "s3://my-bucket/file2.parquet"],
output=f"fd://census/dc_tract",
).run_batch()
To ingest multiple local files, first upload them to S3 with fused.api.upload then specify an array of their S3 paths as the input to ingest.
Row-based ingestion
Standard ingestion is row-based, where the user sets the maximum number of rows per chunk and file.
Each resulting table has one or more files and each file has one or more chunks, which are spatially partitioned. By default, ingestion does a best effort to create the number of files specified by target_num_files (default 20), and the number of rows per file and chunk can be adjusted to meet this number.
job_id = fused.ingest(
input="https://www2.census.gov/geo/tiger/TIGER_RD18/LAYER/TRACT/tl_rd22_11_tract.zip",
explode_geometries=True,
partitioning_method="rows",
partitioning_maximum_per_file=100,
partitioning_maximum_per_chunk=10,
).run_batch()
Area-based ingestion
Fused also supports area-based ingestion, where the number of rows in each partition is determined by the sum of their area.
job_id = fused.ingest(
input="https://www2.census.gov/geo/tiger/TIGER_RD18/LAYER/TRACT/tl_rd22_11_tract.zip",
output=f"fd://census/dc_tract_area",
explode_geometries=True,
partitioning_method="area",
partitioning_maximum_per_file=None,
partitioning_maximum_per_chunk=None,
).run_batch()
Geometry subdivision
Subdivide geometries during ingestion. This keeps operations efficient when geometries have many vertices or span large areas.
job_id = fused.ingest(
input="https://www2.census.gov/geo/tiger/TIGER_RD18/LAYER/TRACT/tl_rd22_11_tract.zip",
output=f"fd://census/dc_tract_geometry",
explode_geometries=True,
partitioning_method="area",
partitioning_maximum_per_file=None,
partitioning_maximum_per_chunk=None,
subdivide_start=0.001,
subdivide_stop=0.0001,
subdivide_method="area",
).run_batch()
Ingest GeoDataFrame
Ingest a GeoDataFrame directly.
job_id = fused.ingest(
input=gdf,
output="s3://sample-bucket/file.parquet",
).run_batch()
Ingest Shapefile
We recommend you .zip your shapefile and ingest it as a single file:
job_id = fused.ingest(
input="s3://my_bucket/my_shapefile.zip",
output="s3://sample-bucket/file.parquet",
).run_batch()
Ingest with a predefined bounding box schema: partitioning_schema_input
Here is an example of an ingestion using an existing partition schema which comes from a previously ingested dataset. This assumes you've already ingested a previous dataset with fused.ingest().
This may be useful if you are analyzing data across a time series and want to keep the bounding boxes consistent throughout your analysis.
@fused.udf
def read_ingested_parquet_udf(path: str = "s3://sample-bucket/ingested_data/first_set/"):
import fused
import pandas as pd
# Built in fused method to reach the `_sample` file and return the bounding boxes of each parquet
df = fused.get_chunks_metadata(path)
# Since we want our `partitioning_schema_input` specified in `ingest()` to be a link to a parquet file containing bounds coordinates, we will save this metadata as a parquet file
partition_schema_path = path + 'boxes.parquet'
df.to_parquet(partition_schema_path)
return partition_schema_path
partition_schema_path = fused.run(read_ingested_parquet_udf)
job_id = fused.ingest(
input="s3://sample-bucket/file.parquet",
output="s3://sample-bucket/ingested_data/second_set/",
partitioning_schema_input=partition_schema_path
).run_batch()
Example: Comparing bounding boxes from ingested AIS data on different days
- First Ingestion
- Second Ingestion
- Second Ingestion with Defined Partition Schema
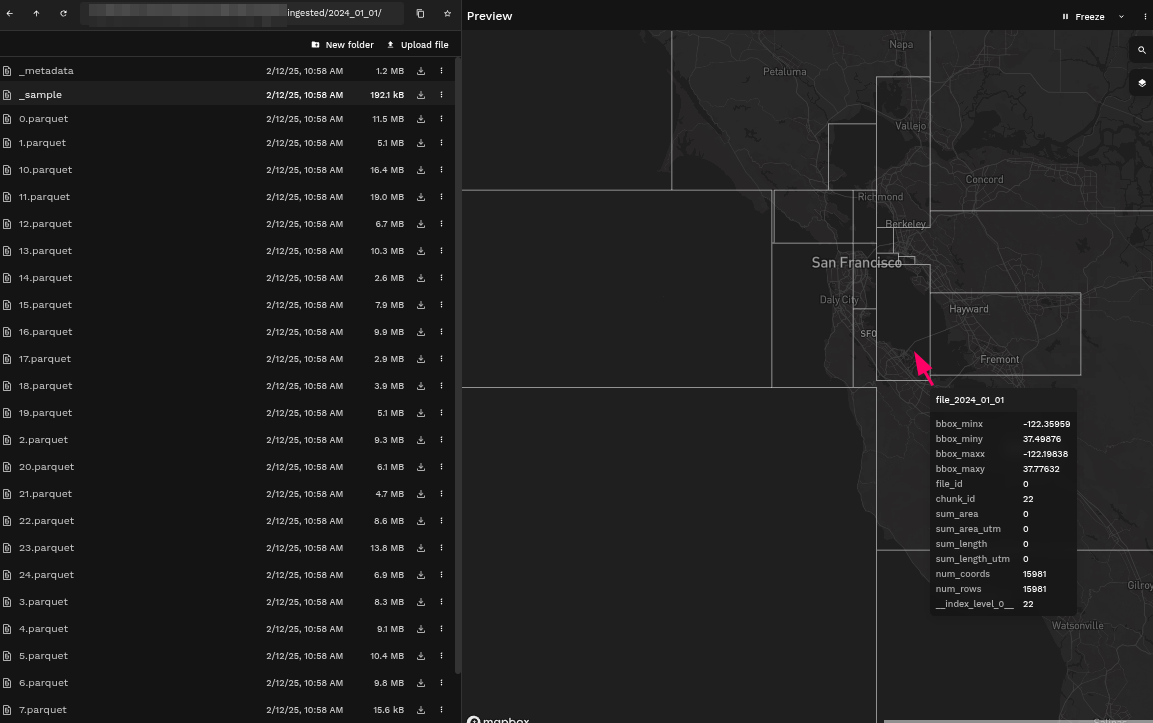
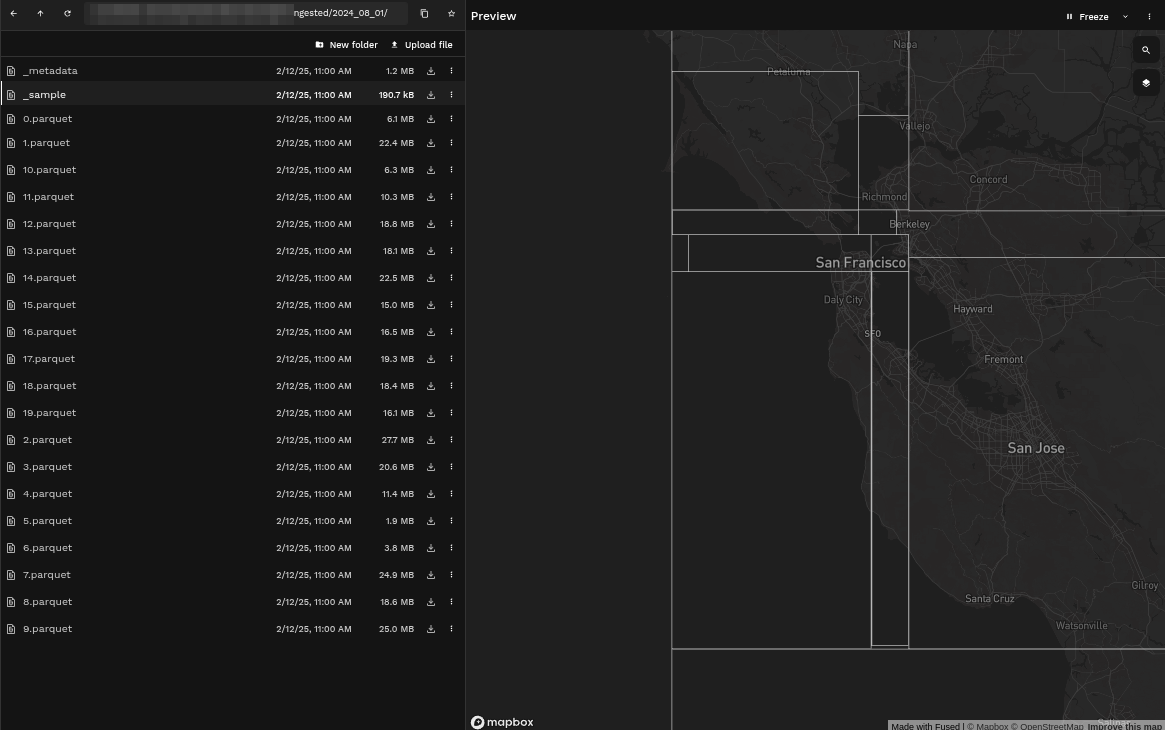
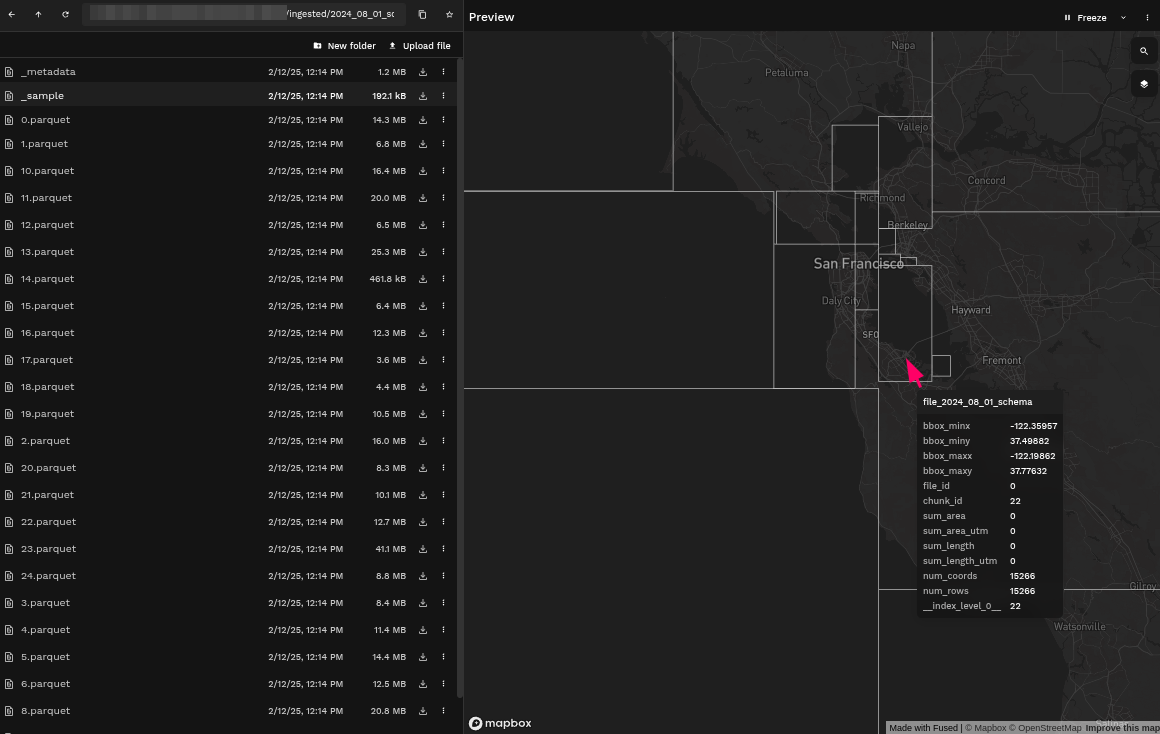
Some of the bounding boxes have changed sizes because of the absence of data in certain areas.
Ingest to your own S3 bucket
fused.ingest also supports writing output to any S3 bucket as long as you have the appropriate permissions:
- Open the
S3 Policytab in the profile page of the workbench and enter your S3 bucket name. This should return a JSON object like this:
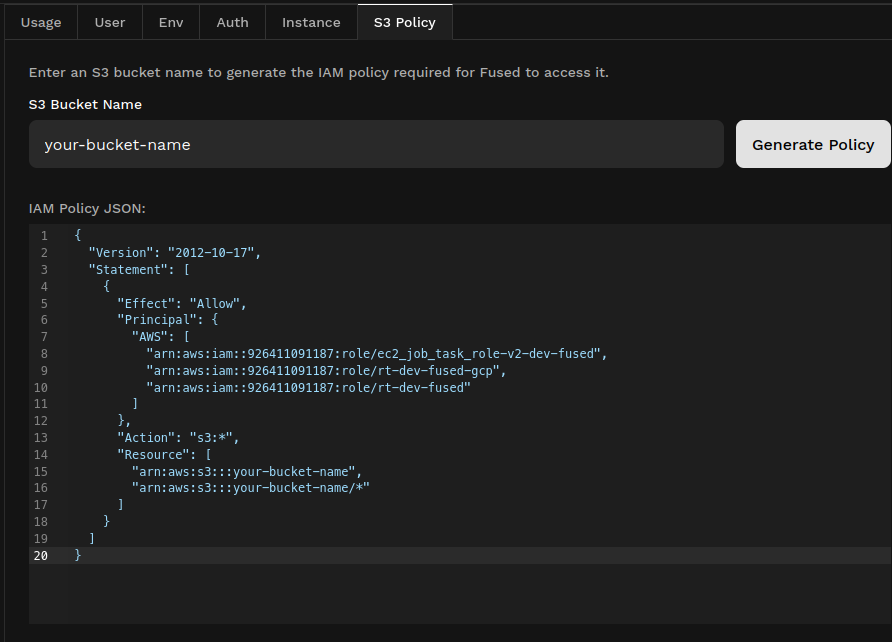
-
Copy this IAM policy and paste it in your AWS S3 Policy tab.
-
Write the output to your S3 bucket using the
outputparameter:
# Assuming s3://my-bucket/ is your own managed S3 bucket
job_id = fused.ingest(
input="s3://my-bucket/file.parquet",
output="s3://my-bucket/ingested/",
).run_batch()
Troubleshooting
As with other Fused batch jobs, ingestion jobs require server allocation for the account that initiates them. If you encounter the following error message, please contact the Fused team to request an increase.
Error: `Quota limit: Number of running instances`